

Models with one of the five simulated covariates influencing clearance and the model without any covariate were fitted to the data. A total of 7400 replicate data sets were simulated independently for each combination of the above conditions. Different magnitudes of residual error and inter-individual variability in the structural model parameters were also introduced to the simulation model. The true covariate influenced clearance according to one of several magnitudes. Data sets, in which each individual had two or three PK observations, were simulated using a one-compartment i.v. The true covariate was set up to have no, low, moderate and high correlation to the other four covariates, respectively. Five covariates were created by sampling from a multivariate standard normal distribution. Data sets with 20-1000 subjects were investigated. The aim of this simulation study was to investigate the effect on power, selection bias and predictive performance of the covariate model, when altering study design and system-related quantities. This can also result in a loss of power to find the true covariates. Competition between multiple covariates may further increase selection bias, especially when there is a moderate to high correlation between the covariates. With such methods, selection bias may be a problem if only statistically significant covariates are accepted into the model.
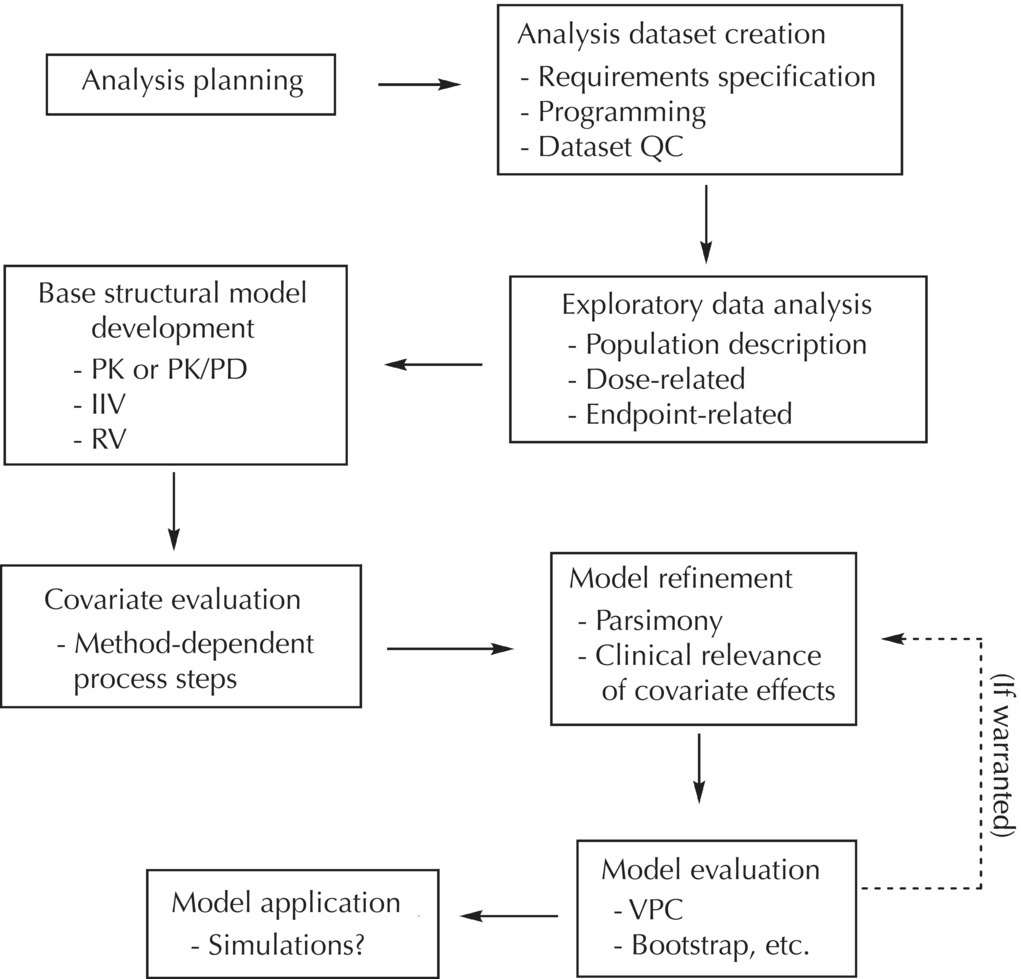
The covariate model is regularly built in a stepwise manner. Identification and quantification of covariate relations is often an important part of population pharmacokinetic/pharmacodynamic (PK/PD) modelling. 109-134 Article in journal (Refereed) Published Abstract Identifiers URN: urn:nbn:se:uu:diva-7923 ISBN: 978-9-3 (print) OAI: oai::uu-7923 DiVA, id: diva2:170387 Public defenceĢ004 (English) In: Journal of Pharmacokinetics and Pharmacodynamics, ISSN 1567-567X, E-ISSN 1573-8744, Vol.

Pharmacokinetics/Pharmacotherapy, Pharmacokinetics, Pharmacodynamics, Modeling, Covariate selection, Stepwise selection, Covariate analysis, Methodology, Model validation, Model evaluation, Type-2 diabetes, Beta-cell function, Meta analysis, Cross-validation, Least absolute shrinkage and selection operator, Pharmacometrics, ED optimization 77 Seriesĭigital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Pharmacy, ISSN 1651-6192 59 Keywords Place, publisher, year, edition, pagesUppsala: Acta Universitatis Upsaliensis, 2007.

The model predictions of beta-cell mass and insulin sensitivity were well in agreement with values in the literature. This model described fasting glucose and insulin levels well, despite heterogeneous patient groups ranging from non-diabetic insulin resistant subjects to patients with advanced diabetes. Alternatively, the performance of SCM can be improved by propagating knowledge or incorporating information from previously analysed studies and by population optimal design.Ī model was also developed on a physiological/mechanistic basis to fit data from three phase II/III studies on the investigational drug, tesaglitazar. In these situations, the lasso method in NONMEM performed better, was faster, and additionally validated the covariate model. Due to selection bias the use of SCM performed poorly when analysing small datasets or rare subgroups. In the course of the investigations, more than one million NONMEM analyses were performed on simulated data. In order to compare the different approaches, investigations were made under varying, replicated conditions. Further, various ways of incorporating information and propagating knowledge from previous studies into an analysis were investigated. The lasso was implemented within NONMEM as an alternative to SCM and is discussed in comparison with that method. The lasso is a penalized estimation method performing covariate selection simultaneously to shrinkage estimation. A further aim was to develop a mechanistic PK-PD model describing fasting plasma glucose, fasting insulin, insulin sensitivity and beta-cell mass. The aim of the current thesis work was to evaluate SCM and to develop alternative approaches. Stepwise covariate modelling (SCM) is commonly used to this end. Identifying covariates which explain differences between patients is important to discover patient subpopulations at risk of sub-therapeutic or toxic effects and for treatment individualization. This is an efficient way of learning about drugs and diseases from data collected in clinical trials. Population pharmacokinetic-pharmacodynamic (PK-PD) models can be fitted using nonlinear mixed effects modelling (NONMEM). 2007 (English) Doctoral thesis, comprehensive summary (Other academic) Abstract


 0 kommentar(er)
0 kommentar(er)
